Structural Assignments to Mycoplasma genitalium (MG) Proteins
SA Teichmann, C Chothia & M Gerstein (1999). "Advances in Structural Genomics," Curr. Opin. Struc. Biol. 9: 390-399.[medline]
Structural Assignments to Mycoplasma genitalium (MG) ProteinsSA Teichmann, C Chothia & M Gerstein (1999). "Advances in Structural Genomics," Curr. Opin. Struc. Biol. 9: 390-399.[medline] |

|
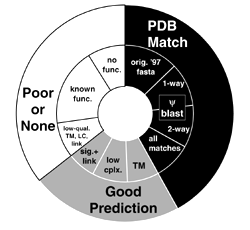
This page provides supplementary related to the above paper. In particular, information is given for the different types of structural assignment that have been made to MG proteins.
For a comparison of the results of other 3D fold assigment to MG of many groups, see: data file (help file).
Two primary sets of MG structural assignments used in the paper:
Standardized assignments of low complexity, transmembrane regions and signal sequences are available from a data directory. In particular:
After the above assignments are performed, some regions of MG remain unassigned.
Groups doing fold assignments: Godzik, Eisenberg, Frishman, Koonin, Bork, Jones, Lengauer, Sali, Levitt, Moult, Sander
![]() TIGR,
the organization that sequenced the genome. The original 468
orf file and new 479
orf file. The MG sequence from NCBI via Entrez.
TIGR,
the organization that sequenced the genome. The original 468
orf file and new 479
orf file. The MG sequence from NCBI via Entrez.