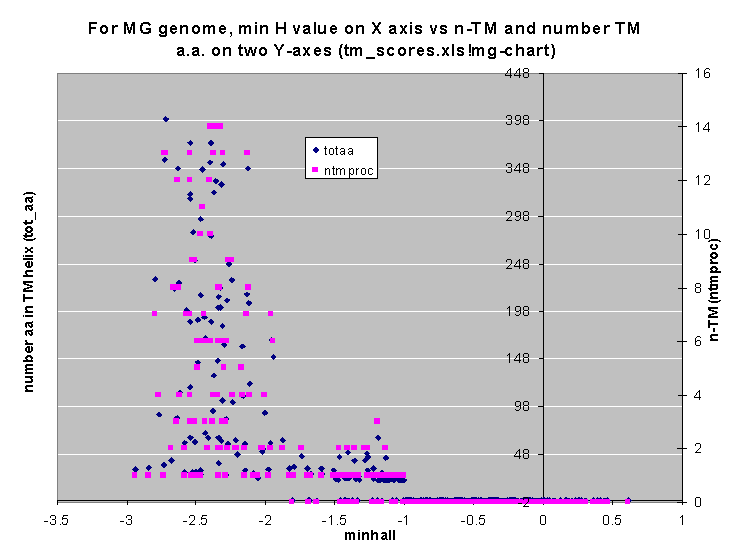
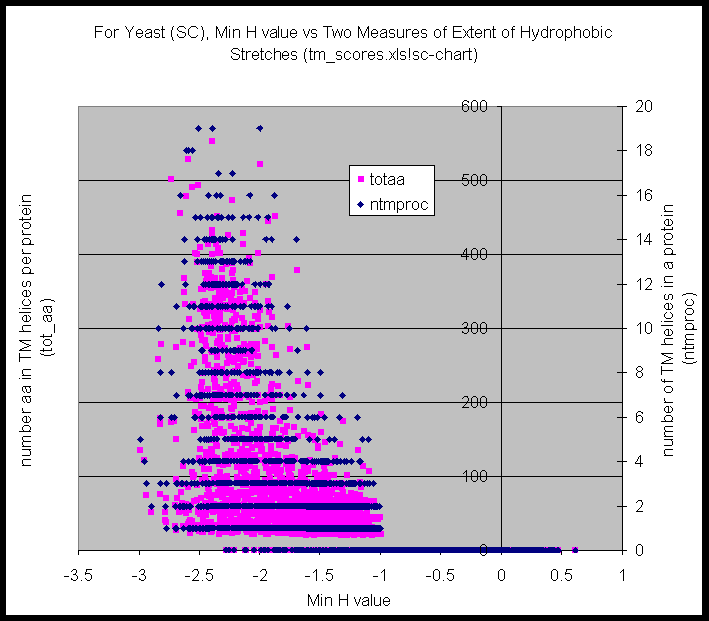
Here are plots Min H value against the number of TM helices per protein (and also number the amino acids in TM helices). There seems to be a clear break around -2 kcal/mole in MG, less so in SC.

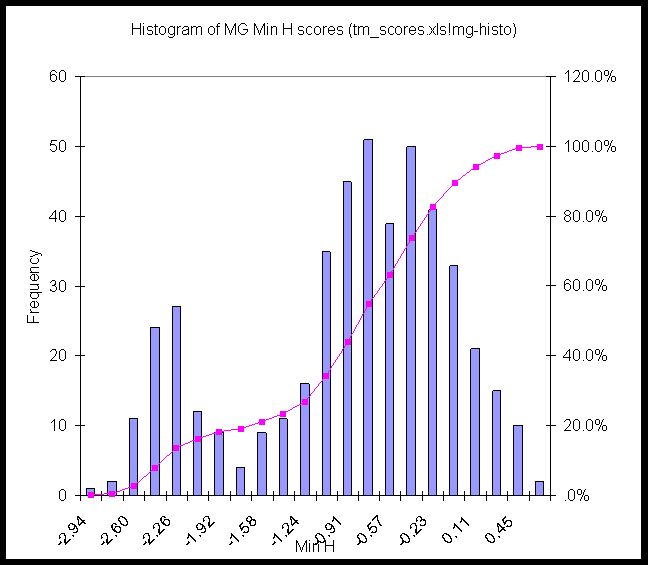
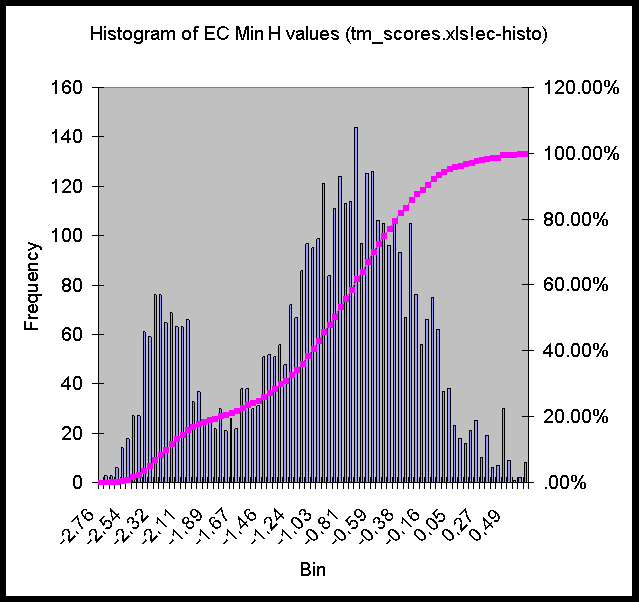
Here are histograms of the Min H values for EC, SC, and MG. There seems to be a clear break around -2 kcal/mole in MG and EC, less so in SC.

Here is the distribution of lengths of low-complexity in SC and MG. There are ~5000 such regions in yeast and ~400 such ones in MG. Notice the similarity in the distribution. This justifies a uniform cutoff of 150 to split low-complexity regions (LCLs) into LCVs and LCMs. LCVs are LCLs longer than 150 aa. LCMs are those shorter.

(This document is derived from genome/tm-lcl-report.doc)