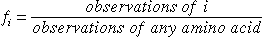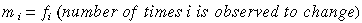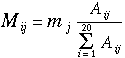Amino Acid Substitution Matrices (continued)
Principles of Scoring Matrix Construction
The Dayhoff Matrix: Proteins evolve through a succesion
of independent point mutations, that are accepted in a population and subsequently
can be observed in the sequence pool.
(Dayhoff, M.O. et al. (1978) Atlas of Protein Sequence and Structure.
Vol. 5, Suppl. 3 National Biomedical Reserach Foundation, Washington D.C.
U.S.A).
First step: Pair Exchange Frequencies
A PAM (Percent Accepted Mutation)
is one accepted point mutation on the path between two sequences, per 100
residues.
Second step: Frequencies of Occurence
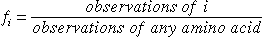 Amino acid frequencies:
Amino acid frequencies:
1978 1991
L 0.085 0.091
A 0.087 0.077
G 0.089 0.074
S 0.070 0.069
V 0.065 0.066
E 0.050 0.062
T 0.058 0.059
K 0.081 0.059
I 0.037 0.053
D 0.047 0.052
R 0.041 0.051
P 0.051 0.051
N 0.040 0.043
Q 0.038 0.041
F 0.040 0.040
Y 0.030 0.032
M 0.015 0.024
H 0.034 0.023
C 0.033 0.020
W 0.010 0.014
Third step: Relative Mutabilities
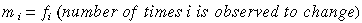 Relative mutabilities of amino acids:
Relative mutabilities of amino acids:
1978 1991
A 100 100
C 20 44
D 106 86
E 102 77
F 41 51
G 49 50
H 66 91
I 96 103
K 56 72
L 40 54
M 94 93
N 134 104
P 56 58
Q 93 84
R 65 83
S 120 117
T 97 107
V 74 98
W 18 25
Y 41 50
All values are taken relative to alanine, which is arbitrarily
set at 100.
Fourth step: Mutation Probability Matrix
The probability that an amino acid in row i
of the matrix will replace the amino acid in column j : the
mutability of amino acid j, multiplied by the pair exchange
frequency for ij divided by the sum of all pair exchange
frequencies for amino acid i:
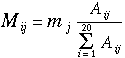
Last step: the log-odds matrix
log to base 10: a value of +1 would mean that the corresponding
pair has been observed 10 times more frequently than expected by chance.
The most commonly used matrix is the matrix from the 1978 edition of the
Dayhoff atlas, at PAM 250: this is also frequently referred to as the MDM78
PAM250 matrix.